Decoding the Genome of Brainea insignis Reveals Insights into Fern Evolution and Conservation
The persistence of rare plant species is not only a matter of individual species survival but also underpins the maintenance of ecosystem functions and the continuity of long evolutionary histories. For monotypic genera, population decline or extinction entails the erosion or complete loss of entire evolutionary lineages, resulting in irreversible consequences for biodiversity and ecosystem stability. Ferns, characterized by large genomes, high chromosome numbers, and widespread aneuploidy and intraspecific polyploid complexity, deviate substantially from classical genetic theories and analytical frameworks that are largely built upon diploid models. Studies relying on second-generation sequencing technologies have traditionally focused on neutral variation in noncoding regions; in the absence of high-quality reference genomes, it remains difficult to distinguish functional coding variation from neutral markers, thereby limiting systematic assessments of natural selection, genetic load, and adaptive evolution. Within the context of conservation genomics, genome-scale investigations integrating genetic drift, inbreeding, and natural selection are therefore urgently needed for ferns.
Research teams led by CHEN Hongfeng and KANG Ming at the South China Botanical Garden, in collaboration with the Shanghai Chenshan Botanical Garden, Shenzhen Fairy Lake Botanical Garden, Nanning Normal University, and Ghent University (Belgium), have systematically elucidated the evolutionary history and genomic basis of endangerment in the rare fern Brainea insignis (Fig. 1). This study reports the first chromosome-level genome assembly of B. insignis, with a total assembly size of 8.62 Gb, and conducts comprehensive comparative and population genomic analyses. The results reveal that B. insignis experienced an ancient whole-genome duplication shared with core leptosporangiate ferns, and that its large genome size is primarily driven by long-term accumulation of long terminal repeat retrotransposons (LTR-RTs), accompanied by an overall slow rate of molecular evolution. Furthermore, the adaptive evolution of its distinctive arborescent stem architecture is closely associated with expansion and strong functional conservation of gene families involved in lignin biosynthesis, providing an explanation for the re-emergence of tree-like growth forms in this evolutionarily distant lineage.

Figure 1. Morphological schematic of Brainea insignis.
Population genomic analyses identified three genetically differentiated lineages from Yunnan (YN), Vietnam (VN), and South China (SC), with the present population structure shaped by multiple historical processes, including Quaternary glaciations, postglacial expansions, and regional gene flow (Fig. 2). Unlike endangered species that have long persisted with extremely small effective population sizes, Brainea insignis has experienced a relatively recent demographic collapse, resulting in a sharp reduction in effective population size, elevated inbreeding levels, and substantial accumulation of deleterious mutations, thereby markedly increasing genetic load. These patterns indicate that purging has not yet become the dominant evolutionary force and that extant populations remain in a trajectory of continued decline. Further environmental association analyses revealed signals of local adaptation correlated with climatic variables across lineages. Genetic offset simulations predict that under future climate change scenarios, the distribution of B. insignis populations will become increasingly fragmented, with populations in the southwestern Indochina Peninsula facing the highest extinction risk (Fig. 3).
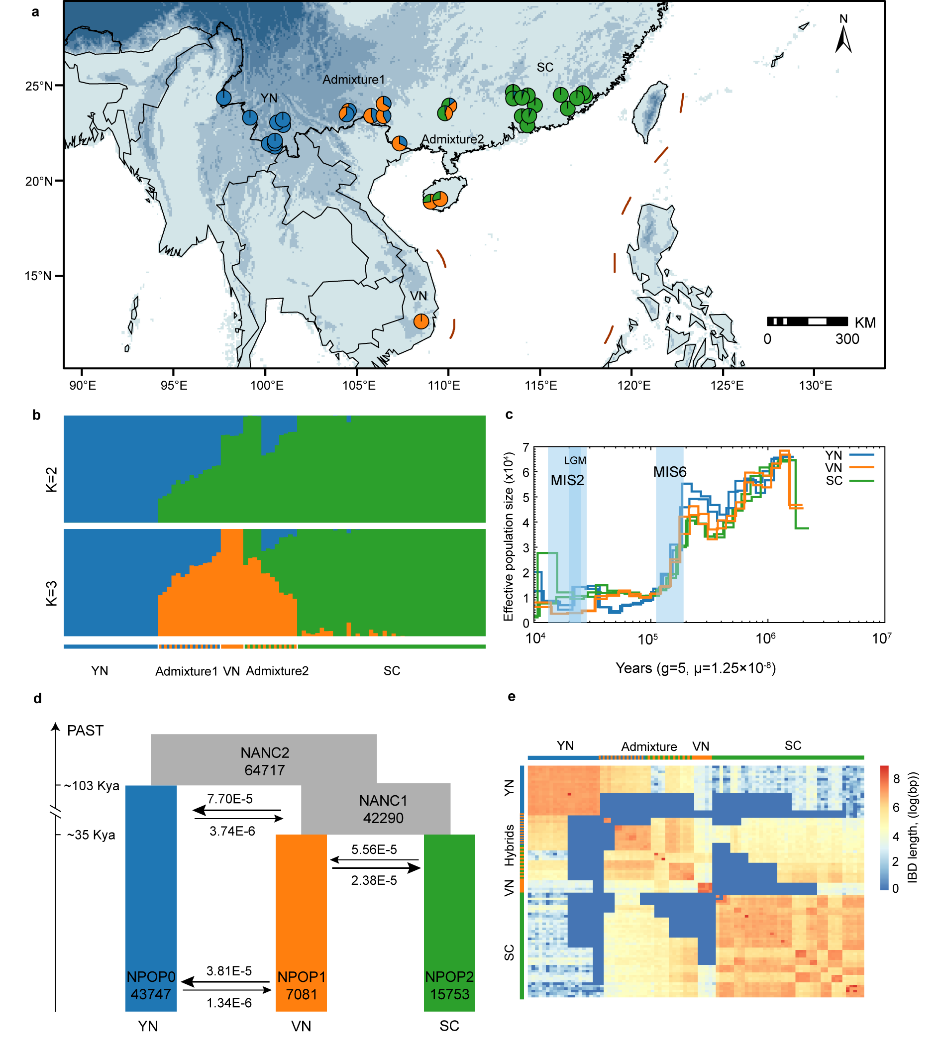
Figure 2. Genetic structure and demographic history of Brainea insig.
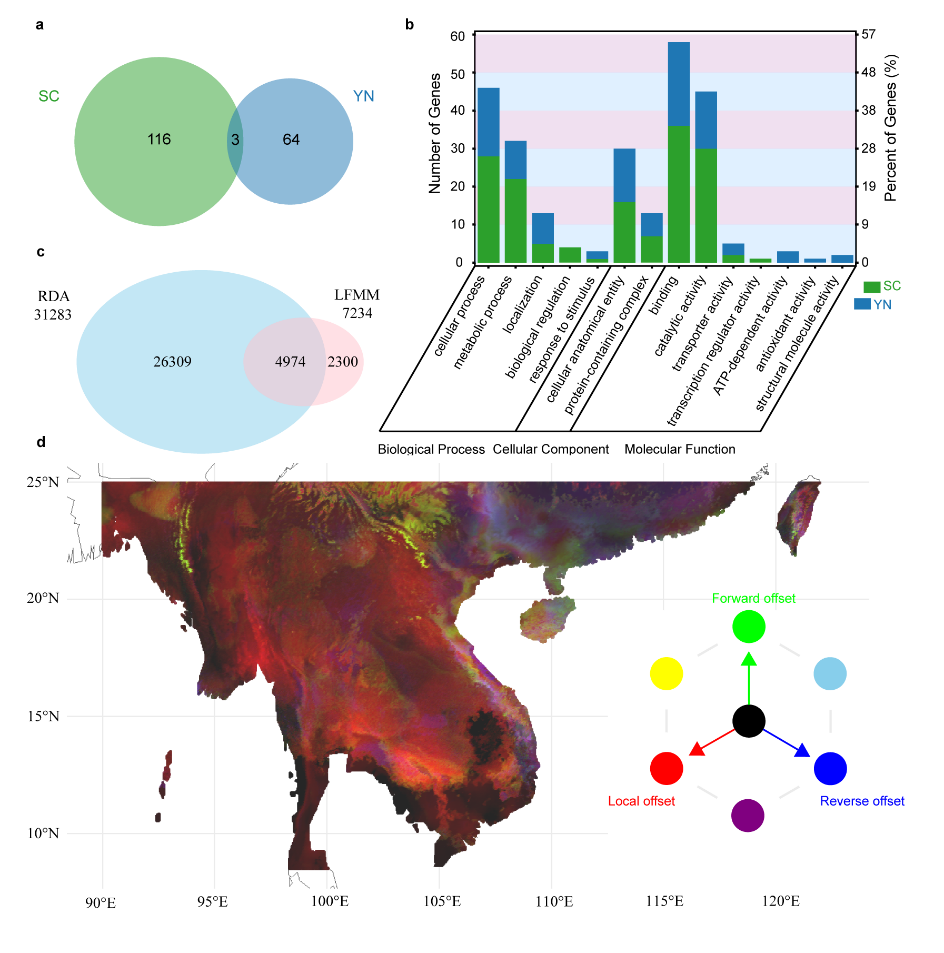
Figure 3. Adaptive differentiation and genetic offset analyses of Brainea insignis.
The study, entitled “Decoding the genome of Brainea insignis reveals insights into fern evolution and conservation,” was published in Nature Communications. This work not only advances fern genomics research and elucidates the pivotal role of demographic history in shaping the genetic architecture of endangered species, but also provides critical theoretical foundations and scientific support for spatially explicit conservation strategies aimed at maintaining habitat connectivity and facilitating adaptive gene flow. XIA Zengqiang, a PhD student at the South China Botanical Garden, is the first author; KANG Ming, WANG Faguo, and Yves Van de Peer are co-corresponding authors. This research was supported by the National Key Research and Development Program of China and the Guangdong Provincial Basic and Applied Basic Research Flagship Program, among others. Article link: https://www.nature.com/articles/s41467-025-68053-0
File Download: