Recent Advances in Pan-genome Analysis of Forest Trees
Understanding the genetic basis of local adaptation is a crucial question in evolutionary biology. Previous studies, relying on single reference genomes and short-read sequencing, have struggled to comprehensively resolve structural variants (SVs) — large-scale genomic alterations proven to regulate stress-response genes and drive phenotypic divergence. With the advent of long-read sequencing technology, graph-based pan-genomes have been developed to identify SVs based on multiple high-quality assemblies. While such approaches have uncovered SV-adaptive associations in model species, pan-genome resources remain limited in non-model long-lived forest trees, preventing this insights into their adaptive evolution.
Oaks (genus Quercus), comprising over 450 species across the Northern Hemisphere, representing an interesting system for studying adaptive evolution. Two East Asian sister species Quercus variabilis and Q. acutissima, which diverged in the late Neogene, span a climatic gradient from temperate to subtropical zones. Long-term sympatric coexistence has facilitated extensive gene flow between them, with population genomic evidence indicating that introgression contributes to adaptive evolution. However, previous studies, constrained by single-reference genomes and short-read data, overlooked the role of SVs in their adaptation.
A research team led by Dr. WANG Baosheng from the South China Botanical Garden (Chinese Academy of Sciences, CAS), in collaboration with the Institute of Botany (CAS), the Research Institute of Forestry (Chinese Academy of Forestry), the Swedish University of Agricultural Sciences, and Umeå University, has constructed the first graph-based pan-genome for Q. variabilis, marking a critical advancement in forest tree genomics. By analyzing 22 individuals, they identified 543,372 high-quality SVs and revealed their roles in climate adaptation. Using whole-genome resequencing data from both Q. variabilis and Q. acutissima, the researchers identified a 250 kb genomic region on chromosome 9 (Chr9-ERF) as a key locus for local adaptation. They further demonstrated that advantageous variants in this region were introgressed from western populations Q. acutissima into Q. variabilis. The study fills a long-standing gap in forest tree pan-genomic resources and provides a framework to dissect rapid adaptation mechanisms in hybridized species, offering actionable insights for climate-resilient forest management.
The research article was published online in Molecular Biology and Evolution titled “Pan-genome analysis reveals local adaptation to climate driven by introgression in oak species”, for details see DOI: 10.1093/molbev/msaf088.
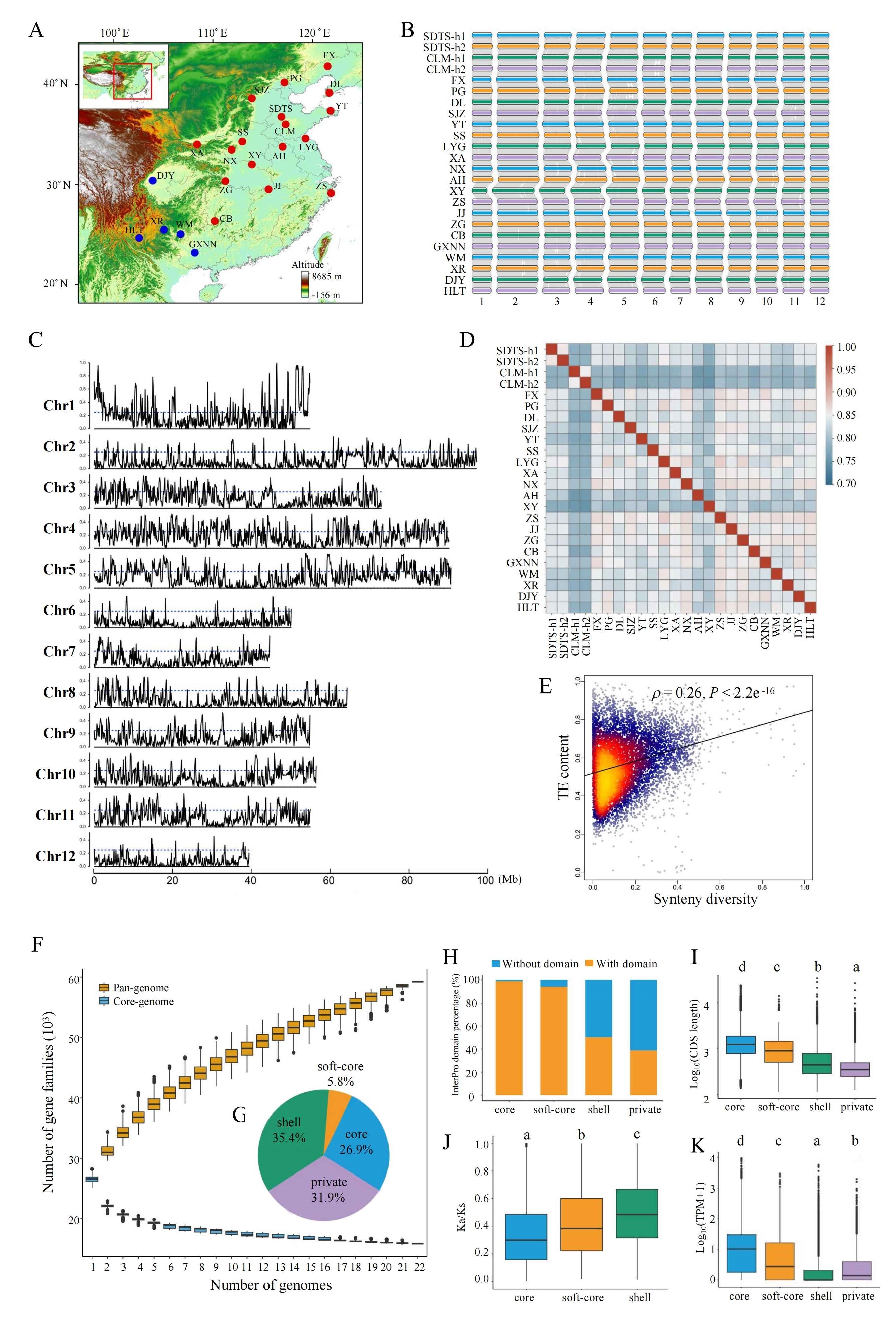
Fig. 1. Genome assembly and pan-genome analyses of 22 Q. variabilis accessions.
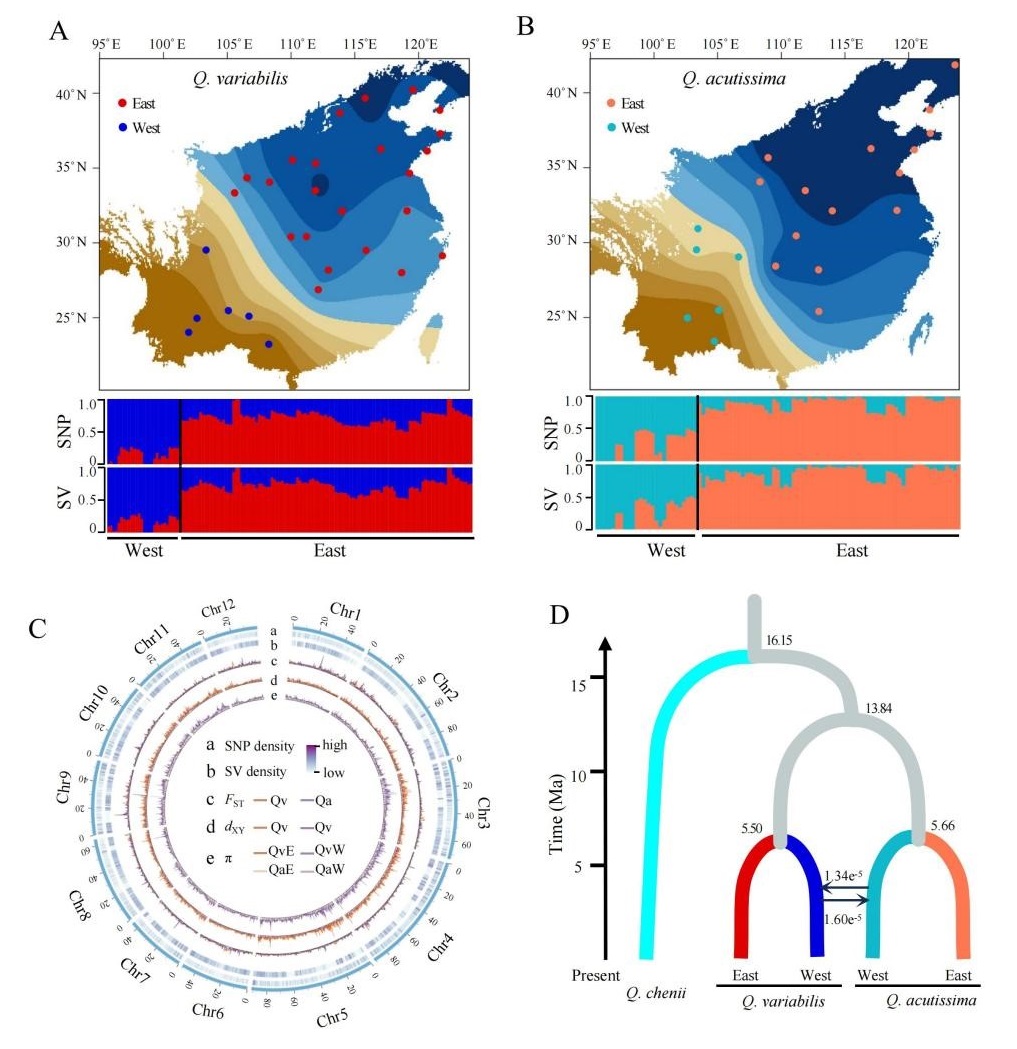
Fig. 2. Genetic divergence and demographic history of Q. variabilis and Q. acutissima.
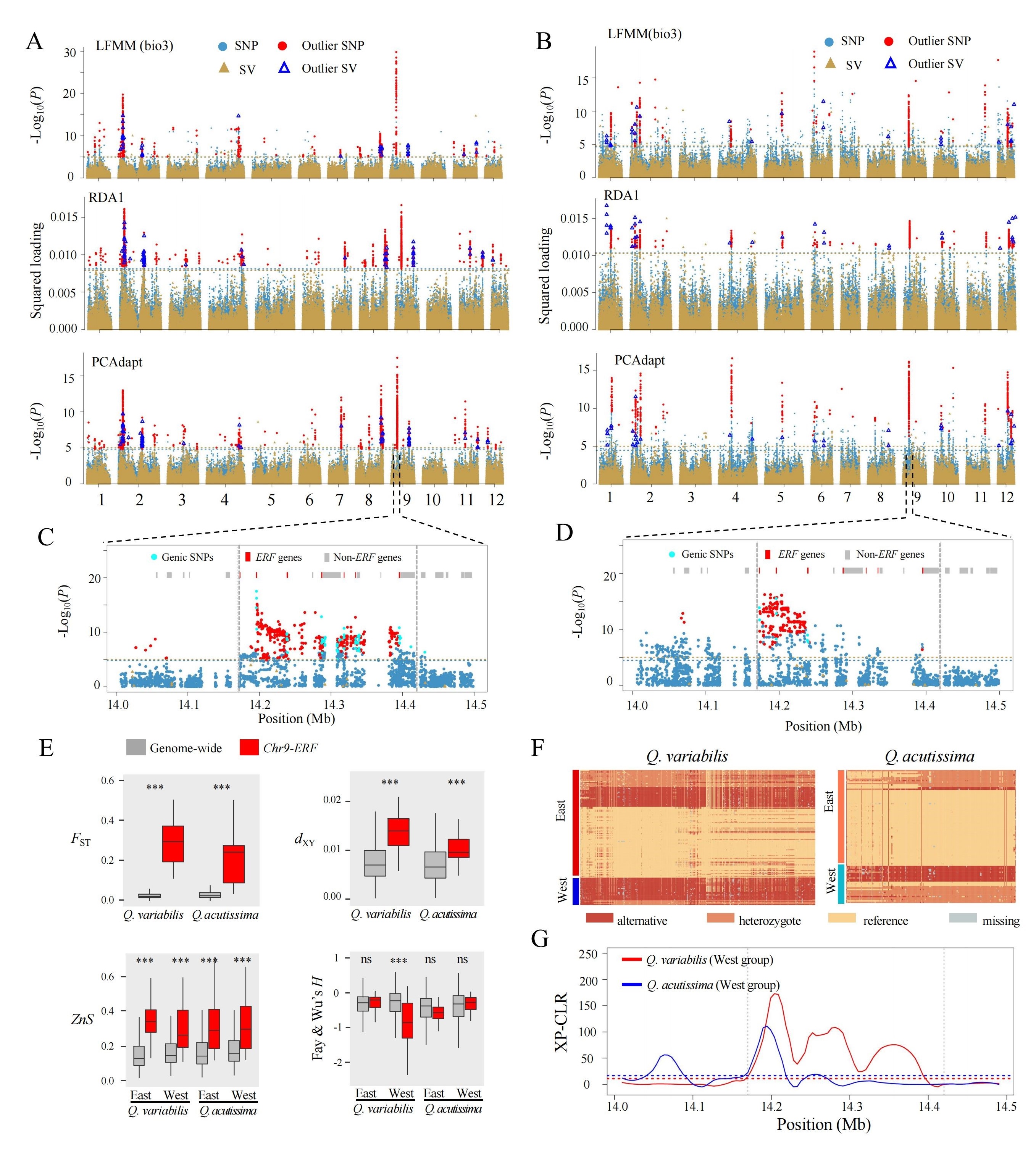
Fig. 3. Genomic signatures of local adaptation in Q. variabilis and Q. acutissima.
File Download: