Genomic Variation Patterns Between Castanopsis Species Revealed
A research team led by Professor Wang Baosheng at the South China Botanical Garden, Chinese Academy of Sciences, has revealed the evolutionary mechanisms driving the similar genomic variation patterns among species of Castanopsis (Fagaceae). This research was supported by the Guangdong Provincial Basic and Applied Basic Research Flagship Project and the Guangdong Provincial Key Laboratory Project.
Understanding the origin and maintenance of genetic variation is a central topic in evolutionary biology. Previous studies have shown that landscapes of genomic variation patterns correlated across different species, which is influenced by multiple evolutionary forces. However, the relative contributions of different mechanisms to the formation of similar genomic variation landscapes, and the complex interactions between these mechanisms, remain unclear. Moreover, the correlated landscape of genomic variation has primarily been studied in recently diverged species, or small numbers of distantly related species, lacking systematic investigation of genomic variation landscapes over longer evolutionary timescales.
This study first assembled a high-quality genome for Castanopsis eyrei. The team then sampled populations of 12 Castanopsis species widely distributed in subtropical evergreen broad-leaved forests. Through whole-genome resequencing, they identified over 50 million single-nucleotide polymorphism (SNP) sites. Population genomic analyses revealed highly correlated genomic variation landscapes across these species. Furthermore, genetic variation strongly correlated with recombination rates and gene density. This suggests that long-term linked selection and conserved genomic features have jointly shaped the common genomic variation landscapes.
Furthermore, by tracking how the correlations between various genetic parameters change with increasing species divergence times, they confirmed that, in addition to background selection, recurrent selective sweeps are also important drivers of the observed patterns of genomic variation. The research also employed a comprehensive analysis of allelic distribution patterns, the accumulation of genetic load, and signals of positive selection across different species' genomes. It revealed the significant role of interspecies introgression, particularly adaptive introgression, in shaping genomic variation landscapes in Castanopsis species, while also uncovering the complex processes and mechanisms driven by multiple evolutionary forces that shape these genomic variation landscapes.
The research article was published online in Molecular Biology and Evolution, titled “Evolution of the correlated genomic variation landscape across a divergence continuum in the genusCastanopsis”. For details see https://doi.org/10.1093/molbev/msae191
Corresponding author: Dr. Baosheng Wang is a scientific researcher at the South China Botanic Garden (CAS). He obtaining PhD from Umea University (Sweden) in 2013, followed by a postdoctoral position in Duke University until 2018. His current research focuses on utilizing population genomic data to investigate the genetic basis of local adaptation in Oak species and predicting how plant species respond to future climate changes.Email: baosheng.wang@scbg.ac.cn
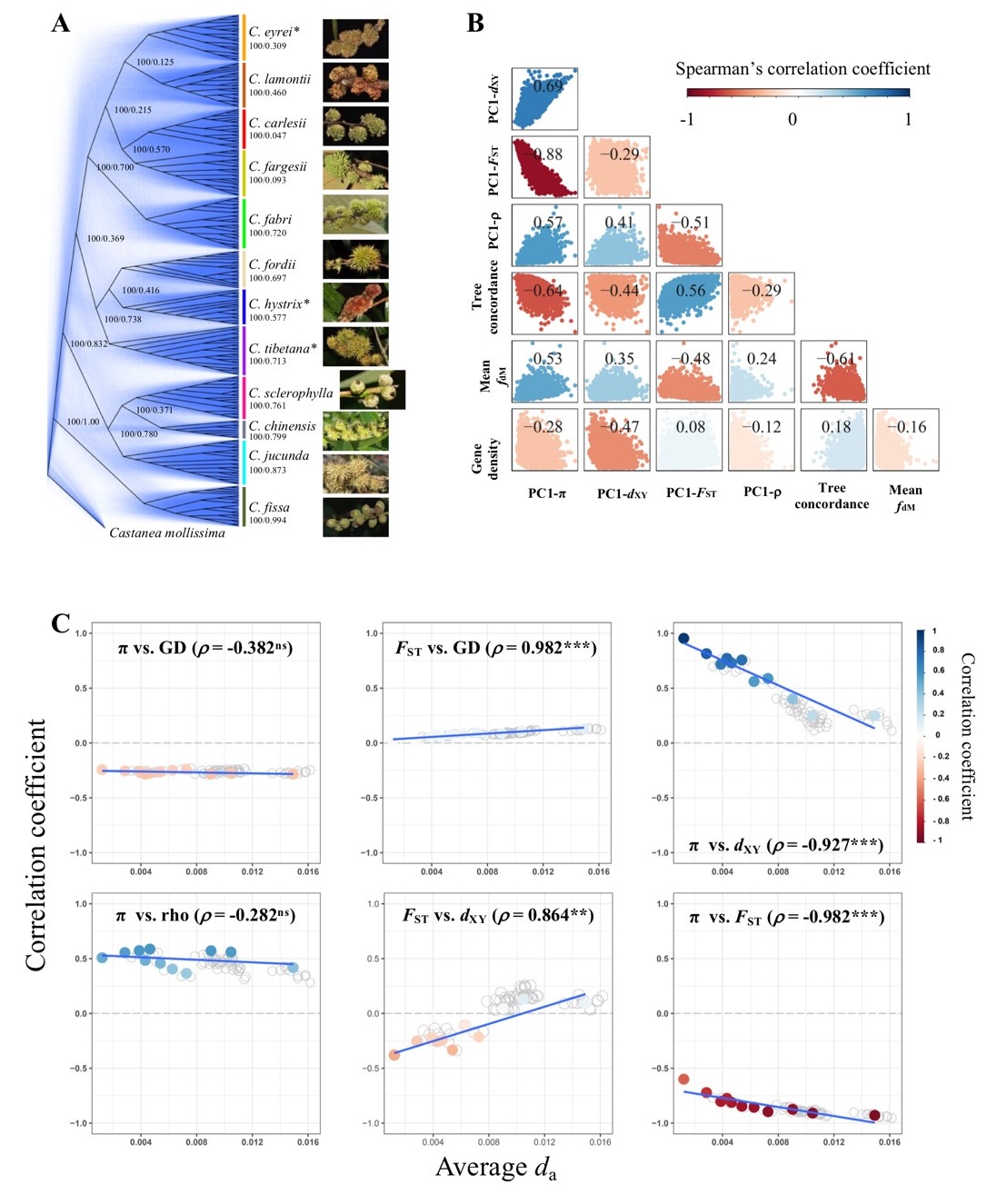
Figure. 1. Mechanisms driving genomic variation in Castanopsis species.
File Download: