The first chromosome-level reference genomes of the ornamental banana and pink banana
Banana (Musa spp.) comprises approximately 70 herbaceous species, which are distributed in tropical and subtropical regions of Asia and Oceania. This genus is renowned for being one of the most important food crops globally and popular ornamental plants in the markets. Although the rapid development of third-generation sequencing technologies has advanced the publishment of high-quality genomes, there is still a lack of genomic resources for banana cultivars, their wild relatives, and ornamental species of Musa, which hampered the crop and ornamental plant improvement.
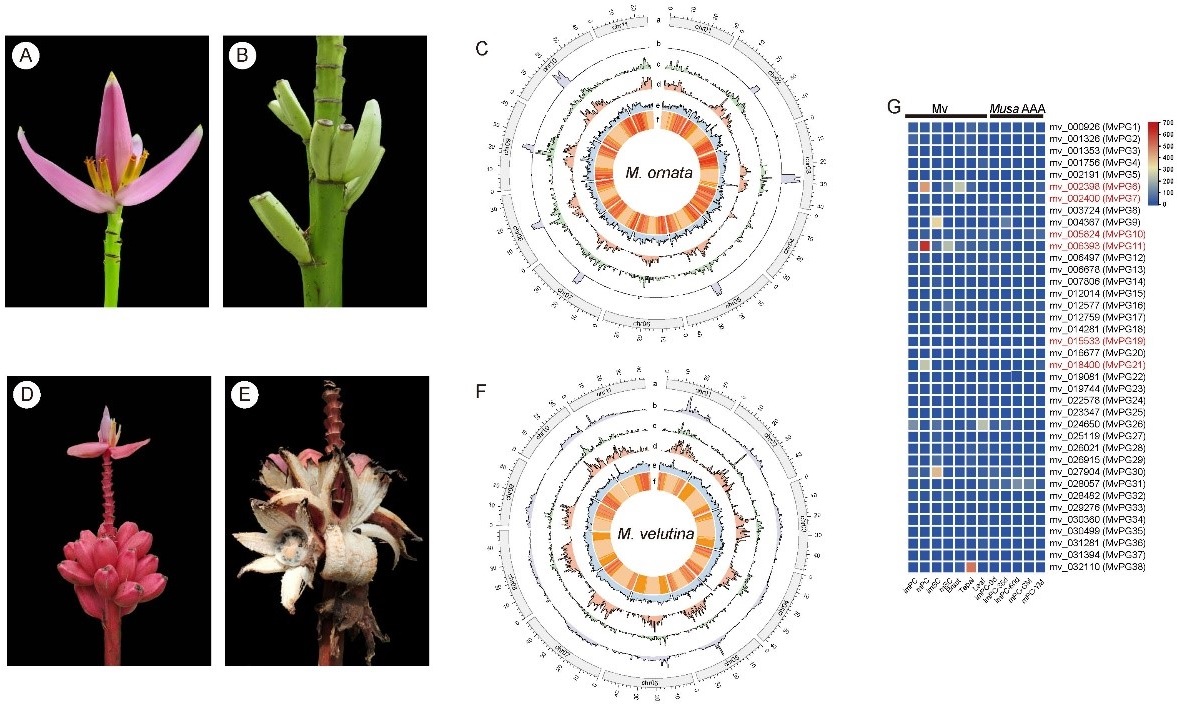
Musa ornata W. Roxburgh (Mo) and Musa velutina H. Wendl. & Drude (Mv) belong to the section Musa of the Musaceae family and are closely related to Musa acuminata. Mo and Mv are native to Bangladesh, Myanmar and northeast India, and are widely cultivated in the tropical countries as popular ornamental plants. In addition, their fruits are also a source of food for the local people. Therefore, Mo and Mv are desirable candidates for generating high-quality genomes to aid future molecular breeding endeavors. In this study, the chromosomal-level genome assemblies of Mo and Mv were generated using Oxford Nanopore long reads and Hi-C reads. The genomes of Mo and Mv were assembled into 11 pseudochromosomes with genome sizes of 427.85 Mb and 478.10 Mb, respectively. Repetitive sequences comprised 46.70% and 50.91% of the total genomes of Mo and Mv, respectively. Genome quality assessments confirmed the contiguity (LAI: 13.68 and 16.81), accuracy (mapping rates of Illumina reads: 95.55% and 94.29%), and completeness (BUSCO: 98.08% and 98.51%) of the two genomes. According to the gene predictions, a total of 39,177 and 31,256 high-confidence protein-coding genes were annotated for Mo and Mv, respectively. Compared to Musa acuminata and Mv, several large inversions and translocations were detected on chr04 of Mo. Differentially expressed gene (DEG) and Gene Ontology (GO) enrichment analyses indicated that upregulated genes in the mature pericarps of Mv were mainly associated with the saccharide metabolic processes, particularly at the cell wall and extracellular region. Furthermore, several polygalacturonase (PG) genes were identified that showed higher expression levels in mature pericarps of Mv compared to other tissues, which may be accountable for pericarp dehiscence. In addition, this study also identified genes associated with anthocyanin biosynthesis pathway. Taken together, the chromosome-level genome assemblies of Mo and Mv provide valuable insights into the mechanism of pericarp dehiscence and anthocyanin biosynthesis in banana, which will significantly contribute to future genetic and molecular breeding efforts.
The article “Chromosome-level genome assemblies of Musa ornata and Musa velutina provide insights into pericarp dehiscence and anthocyanin biosynthesis in banana” has been published in Horticulture Research. This work was supported by the National Natural Science Foundation of China. For further information, please refer to: https://academic.oup.com/hr/advance-article/doi/10.1093/hr/uhae079/7628330
First author information: Tian-Wen Xiao, Post doctor, Key Laboratory of National Forestry and Grassland Administration on Plant Conservation and Utilization in Southern China, South China Botanical Garden, Chinese Academy of Sciences. Research field: Plant systematics and evolution. Email: xiaotianwen@scbg.ac.cn
File Download: