Shifts in fungal biomass and activities of hydrolase and oxidative enzymes explain different responses of litter decomposition to nitrogen addition
Exogenous nitrogen (N) input is a key factor affecting litter decomposition. Yet our mechanistic understanding of microbial community and EE response to N addition and their associated effect on litter decomposition is still limited.
In a research article recently published in the journal Biology and Fertility of Soils, researchers from South China Botanical Garden of Chinese Academy of Sciences conducted a field experiment to examined how simulate N deposition to influence litter decomposition by combining the microbial community properties with principles of enzyme kinetic and thermodynamic. They found N addition significantly enhanced litter decomposition at the initial stage of litter decomposition by stimulating enzyme-substrate affinity (Km) and maximum velocity (Vmax) of enzyme involved in C and N mineralization, likely as a result of alleviating N limitation of microbial community. However, these effects did not persist into later decomposition stages. Instead, N addition slowed decomposition as kinetic parameters (Vmax, Km, and Vmax/Km) declined and fungal abundance decreased. However, N addition did not alter the activation energy (Ea) of all enzyme.
Their study identifies the mechanisms that contribute to the N effect has on litter decomposition. Specifically, we found that a decrease in fungal abundance and the catalytic efficiency of C, N, and P-acquiring hydrolytic enzymes have the greatest potential to lead to decrease in forest litter decomposition, with increasing N addition. They provide evidence of microbial communities as drivers of EE activity response to high N deposition through mediating enzymatic kinetic characteristics. Their findings provide a novel experimental contribution to the understanding of the relationship between EE kinetics, microbial communities and substrates availability. The analyses also show that Vmax/Km should be incorporated into enzyme based model due to decoupling of Vmax and Km and it is sensitive to variation in controlling factors.
For further reading, please refer to: http://link.springer.com/10.1007/s00374-020-01434-3.
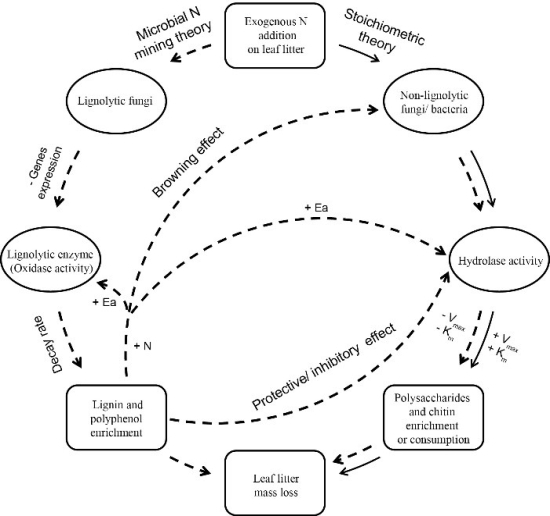
Figure. Conceptual diagram of microbial community and enzyme responses to N addition.
File Download: