Histone replacement during chromatin-remodeling plays a key role in transcriptional regulation. Replacing their canonical counterparts by histone variants often leads to gene expression changes in eukaryotes, which is one of the most important biological responses to stresses caused by temperature, drought, salinity and heavy metals.
Dr. David W. OW’s group had reported previously that overexpression of a plant-specific protein, Arabidopsis OXIDATIVE SRESS3 (AtOXS3), was able to enhance stress tolerance in both Arabidopsis and Schizosaccharomyces pombe. Since AtOXS3 has a putative N-acetyltransferase catalytic domain which was critical for stress tolerance and was found co-localized with histone H4, AtOXS3 was speculated to be a chromatin-associated factor which could regulate the expression of certain resistance genes by remodeling chromatin. Ph.D. student LAI Dingwang and colleagues used a fission yeast (S. pombe) model system, which has been long served for studying oxidative stress responses, to probe into the mechanism of AtOXS3 enhancing stress tolerance and tried to shed light on how this protein might operate in its native host, Arabidopsis.
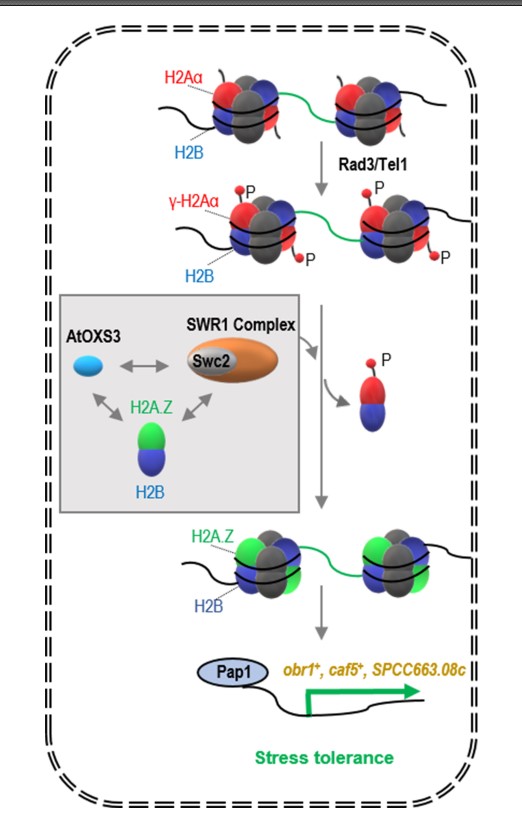
Based on the experimental results on S. pombe, the following working model of ‘AtOXS3-mediated stress tolerance’ was proposed (Figure 1). Under abiotic stress, the histone H2Aα deposited in drug-resistant gene promoters is phosphorylated by Tel1/Rad3 kinases, which leads to the eviction of H2Aα from chromatin. AtOXS3 subsequently causes the replacement of H2Aα with histone H2A.Z, by interacting with H2A.Z and the Swc2 subunit of the chromatin remodeling complex SWR1. Finally, the oxidative stress-responsive transcription factor Pap1 is recruited to the promoters of the drug-resistant genes to activate their expression and the cells are primed for higher tolerance to the stresses of at least diamide and Cd. Although how AtOXS3 operates in its native host has yet to be investigated, this study has unraveled a probable mechanism on how a heterologous protein reorganizes the chromatin landscape to specific genes for transcription activation and enhances host tolerance.
This study was conducted by Ph.D. student LAI Dingwang (first author) and colleagues under the instructions by the corresponding authors, Dr. David W. OW and Dr. WANG Changhu. The relevant paper entitled “Arabidopsis OXIDATIVE STRESS 3 enhances stress tolerance in Schizosaccharomyces pombe by promoting histone subunit replacement that upregulates drug resistant genes” has been published in “Genetics (2021), iyab149 (IF: 4.562)”. This work was supported by the National Key Research & Development Project from the Ministry of Science and Technology of China, and by the Key Research Program of Frontier Sciences, Chinese Academy of Sciences. For further reading, please refer to https://doi.org/10.1093/genetics/iyab149